We use physiology to link genomic variation to organismal fitness in order to understand...
how physiology evolves to fit organisms to their ecologies, &
how evolution shapes genetic and biochemical pathways underlying physiological change.
The pathways of physiology provide systems of genes that link genetic variation and divergence to whole-organism physiological performance traits, such as development rate, metabolic rate, flight velocity, ethanol tolerance and stress responses. These pathways provide our framework for taking an integrative approach to linking genes to their evolutionarily and ecologically significant function.
how physiology evolves to fit organisms to their ecologies, &
how evolution shapes genetic and biochemical pathways underlying physiological change.
The pathways of physiology provide systems of genes that link genetic variation and divergence to whole-organism physiological performance traits, such as development rate, metabolic rate, flight velocity, ethanol tolerance and stress responses. These pathways provide our framework for taking an integrative approach to linking genes to their evolutionarily and ecologically significant function.
Cellular & physiological adaptations to a variable environment
|
The tremendous diversity in form and function across the Tree of Life is matched by an equally amazing diversity of cellular, physiological and behavioral adaptations that enable organisms to function in their environments. We investigate the genetic and energetic bases for plastic and fixed adaptive strategies - including how cellular and physiological plasticity evolves, how embryos defend themselves against thermal stress and how mom’s preferences and her mRNAs might buffer this stress (in collaboration with Brent Lockwood), and how pathways and physiologies that mediate abiotic (e.g., temperature and ethanol) and biotic factors (e.g., pathogens) diverge among natural populations and species of Drosophila. Current collaborative projects include investigations of physiological plasticity in migrating butterflies with the Kroforst Lab at the University of Chicago, and the effects of winter warming on the diapausing stage of the small white cabbage butterfly with Emily Mikucki and Brent Lockwood at the University of Vermont.
|
Membranes, metabolism, temperature & toxins
|
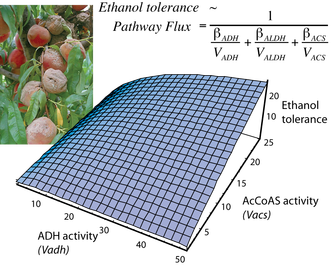
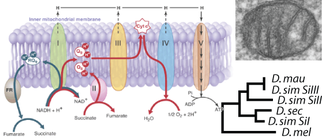
Organisms in nature do not experience single, isolated selection pressures. A remaining challenge is to describe phenotypic evolution as the integrated outcome of multiple selection pressures that may act differently across life stages. We are investigating the cell membrane as an integrator of and responder to the combined effects of environmental ethanol and temperature. Ethanol disrupts cellular function by making membranes more fluid. In addition, because flies are ectotherms, their survival depends upon adjusting membrane fluidity in response to temperature -- a physiological plasticity that we have shown evolves in response to environmental heterogeneity in Drosophila. Aerobic metabolism depends on the mitochondrial membranes that harbor the oxidative phosphorylation and ATP generating protein machinery. We are currently testing whether ethanol tolerance is conferred by the membrane plasticity that maintains respiration in the presence of environmental gradients in ethanol and temperature.
|
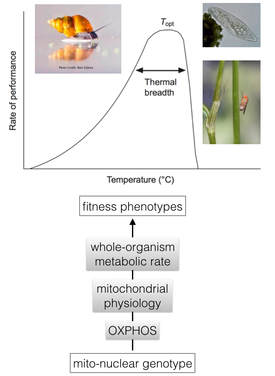
Mitochondrial-nuclear (co)evolution
|
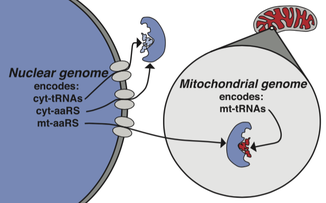
The unique genetics of the mtDNA - namely its putative propensity to accumulate deleterious mutations - are hypothesized to drive compensatory and positive coevolution in nuclear genes. We investigate this important evolutionary dynamic using population genomic and molecular evolutionary analyses, as well as functional approaches to map intergenomic mito-nuclear interactions. We also quantify the functional and evolutionary consequences of this dynamic.
Current collaborative projects include investigations of mito-nuclear effects on female fecundity with the Calvi Lab (Indiana University) and on temperature-dependent male sterility with the Meiklejohn Lab (UNL). We also work with Maurine Neiman (U Iowa) & Joel Sharbrough (Colorado State) to test for genomic and functional signatures of elevated rates of mito-nuclear coevolution in the asexual aquatic snail Potamopyrgus antipodarum. |
Evolutionary & developmental genetics of energetics
|
How do the pathways underlying largely homeostatic and conserved physiological systems nevertheless harbor variation within populations, diverge among populations and evolve across phylogenies?
Energetic traits, such as ATP levels, must be maintained as organisms adapt to different ecologies and dynamically respond to variable environments within their lifetime. These energetic traits also underlie basic processes of cell division, growth and development. We are increasingly interested in the genes and physiologies that link temperature to metabolic rates and growth rates in insects, as well as the developmental trajectory and plasticity of metabolic processes. Our recent work has shown that even the basic scaling laws that govern the relationship between mass and metabolic rate change across development, thermal environments and genotypes. We seek to understand the energetic basis for this. Current collaborative projects include applying models of thermal performance curves to data that integrate across levels of metabolic physiology from enzymes to population-level processes as part of the NSF Rules of Life initiative with John DeLong (UNL). |
Environment, nutrition, hosts and pathogens
|
We have found that compromised mitochondrial function decreases the ability of organisms to survive infection with natural pathogens and generates tradeoffs between immunity and fecundity. Current work in the lab investigates dynamic changes in metabolic rate and feeding behavior during an immune response, and the effects of nutrient resources on the outcomes of infection. With Drs. Jessica Hite and Clay Cressler (UNL), Justin Buchanan and undergraduate Lauren Reiman are investigating whether host and pathogen's nutrient interests are aligned or in conflict, and how this effects the ecological and evolutionary dynamics of disease.
|
Proudly powered by Weebly